A Virtual PCR Simulator for Education
DNA Education Posted 22 August 2025

Try out our educational simulator of the Polymerase Chain Reaction!
Students can tweak 11 different PCR parameters such as the temperature cycle, or the amount of polymerase enzyme added to the reaction, and observe how this impacts the final yield and purity of amplified DNA.
And, all of this happens without using expensive re-agents in the lab! The simulation calculates DNA yield by running an actual (but highly simplified) chemical kinetics model of the DNA amplification process.
The PCR simulator was initially used at Newcastle University UK to teach Design of Experiments methodology (see our paper below). Since then, it has been used in other universities around the world for various projects/classes, and particularly during the covid pandemic. We are always interested to learn how the PCR simulator is being used!
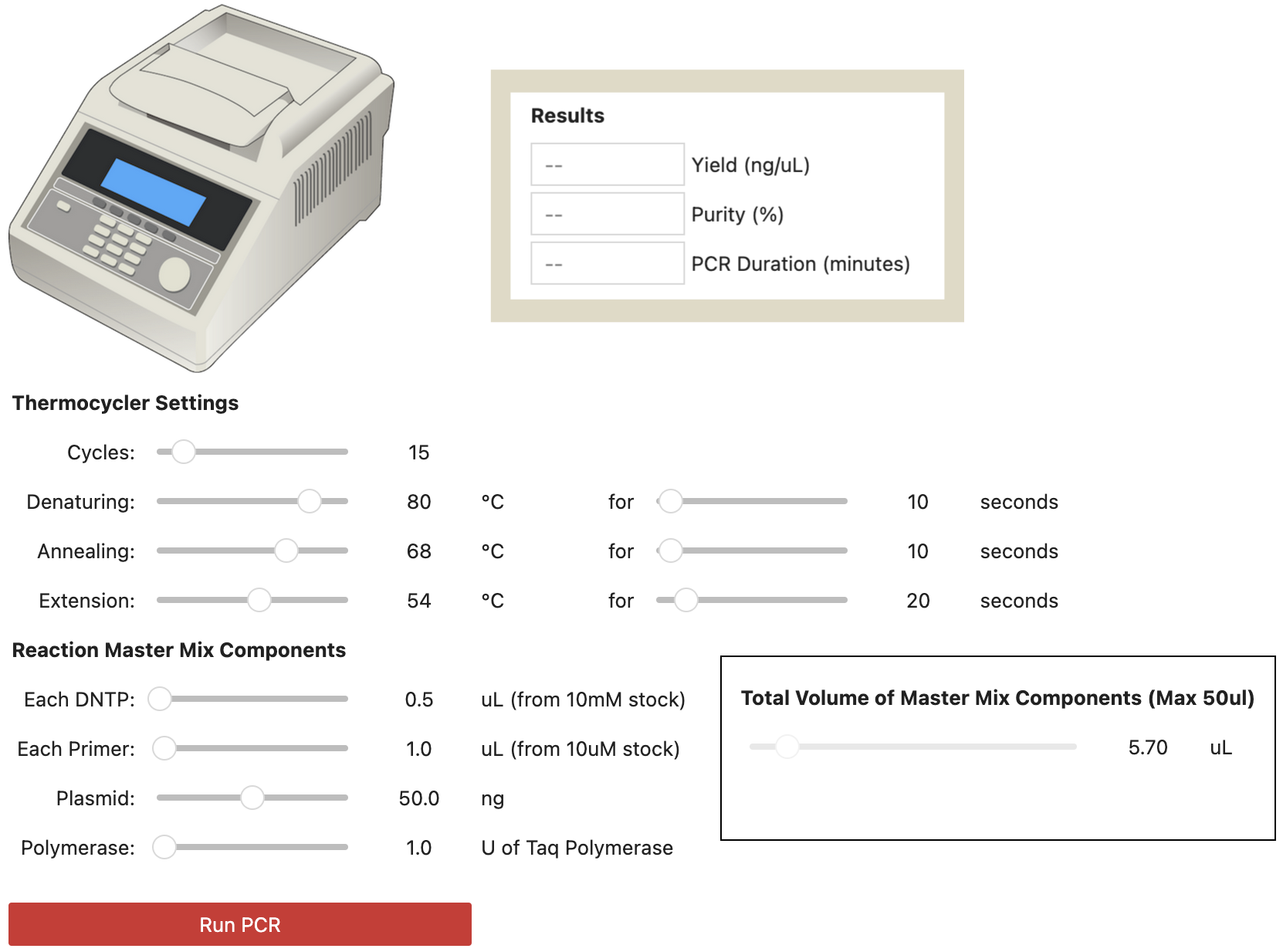
Option 1: Run PCR Simulator Locally on Your Computer
You can run the PCR simulator as a Python Jupyter Notebook on your computer. Download the Git repository https://bitbucket.org/ben_s_e/virtual-pcr-notebook and see the README.txt for how to install and use.
Option 2: Run PCR Simulator on the Web
https://virtual-pcr.ico2s.org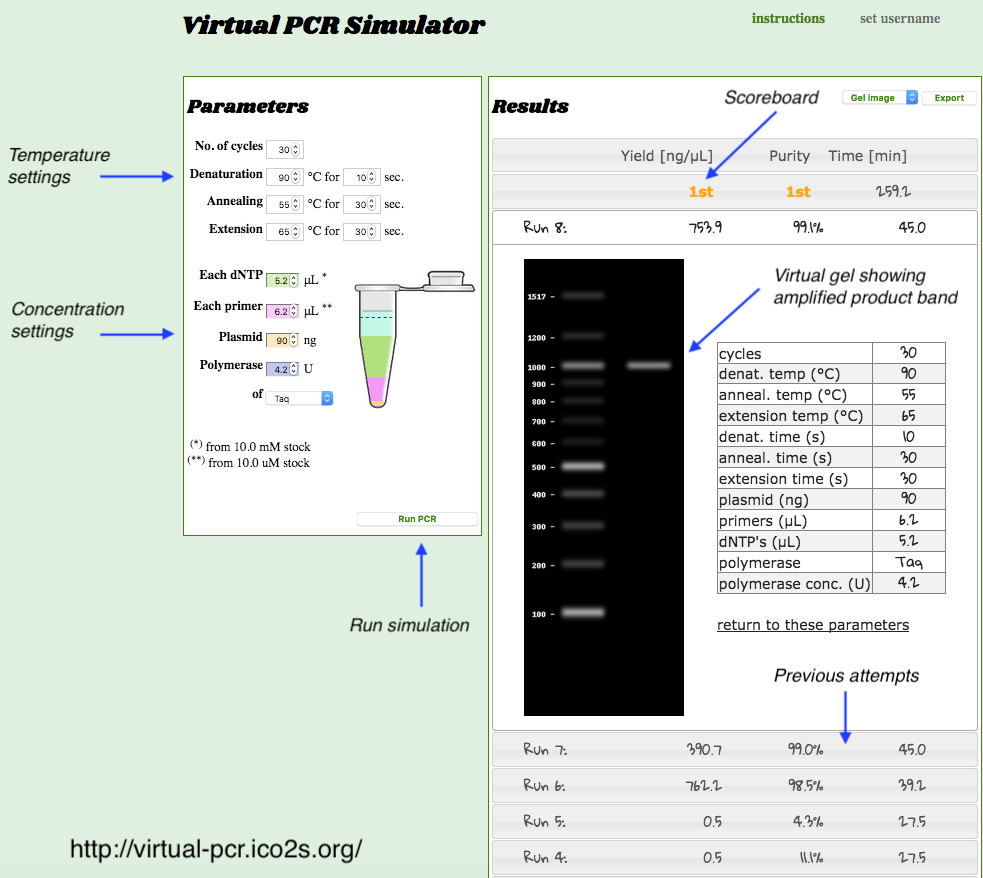
Journal Papers
Fellermann and Shirt-Ediss et al. (2019). Design of experiments and the virtual PCR simulator: An online game for pharmaceutical scientists and biotechnologists. Pharmaceutical Statistics 18(4) 402-406.
Fellermann and Shirt-Ediss et al. (2018). The PCR Simulator: an on-line application for teaching Design of Experiments and the polymerase chain reaction. BioRxiv preprint.
Technical Details
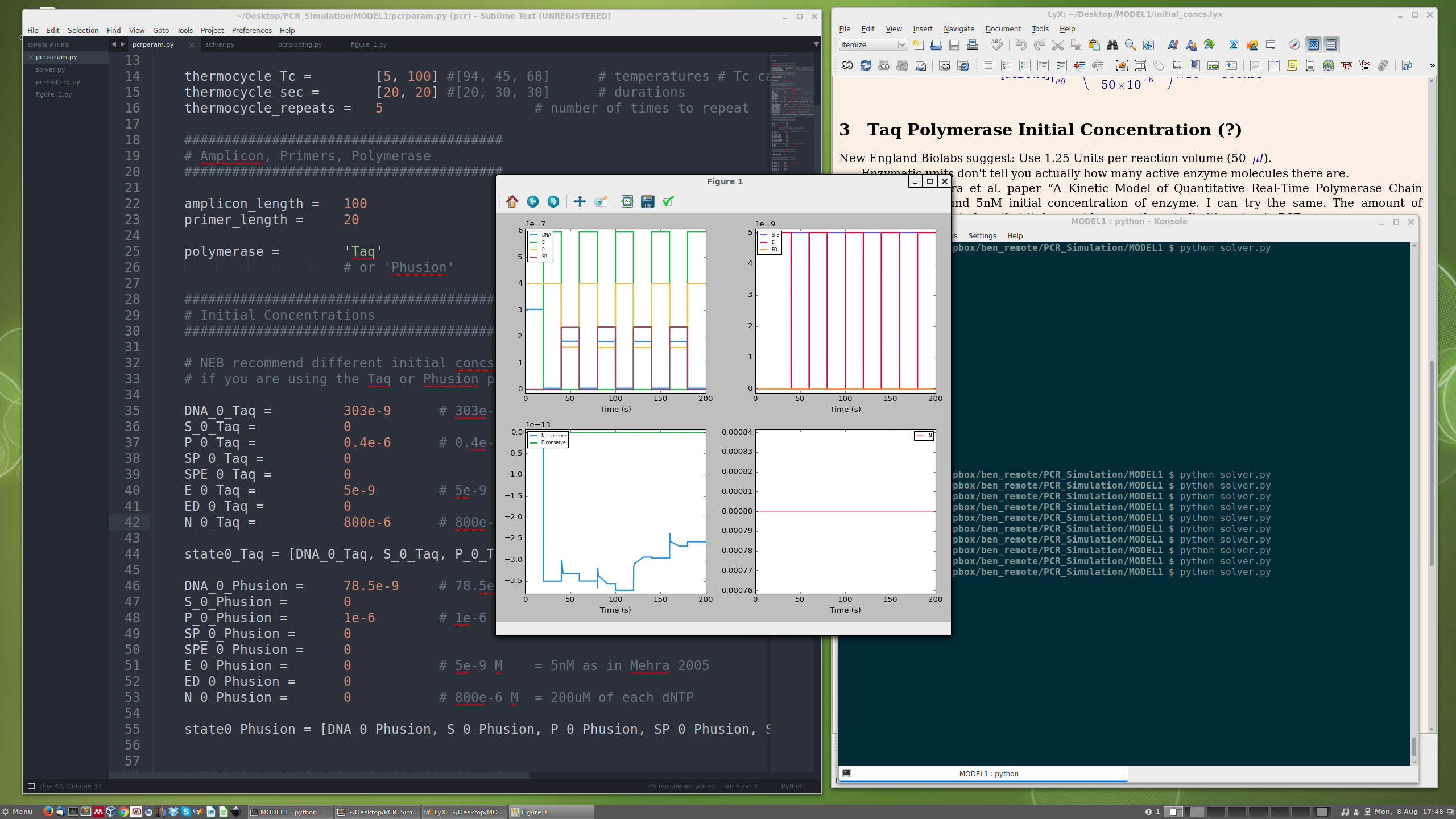
⬆️ Screenshot of PCR Model v1 under development!
Techniques Used
DNA chemical kinetics modelling, DNA thermodynamics, automatic reaction enumeration, ordinary differential equations, model parameterisation, ipython/web user interface development, Python
This work was developed while I was a postdoctoral researcher at Newcastle University, UK.



