Selecting Sequences for DNA Origami Nanotechnology
DNA Nanotech Posted 21 August 2025
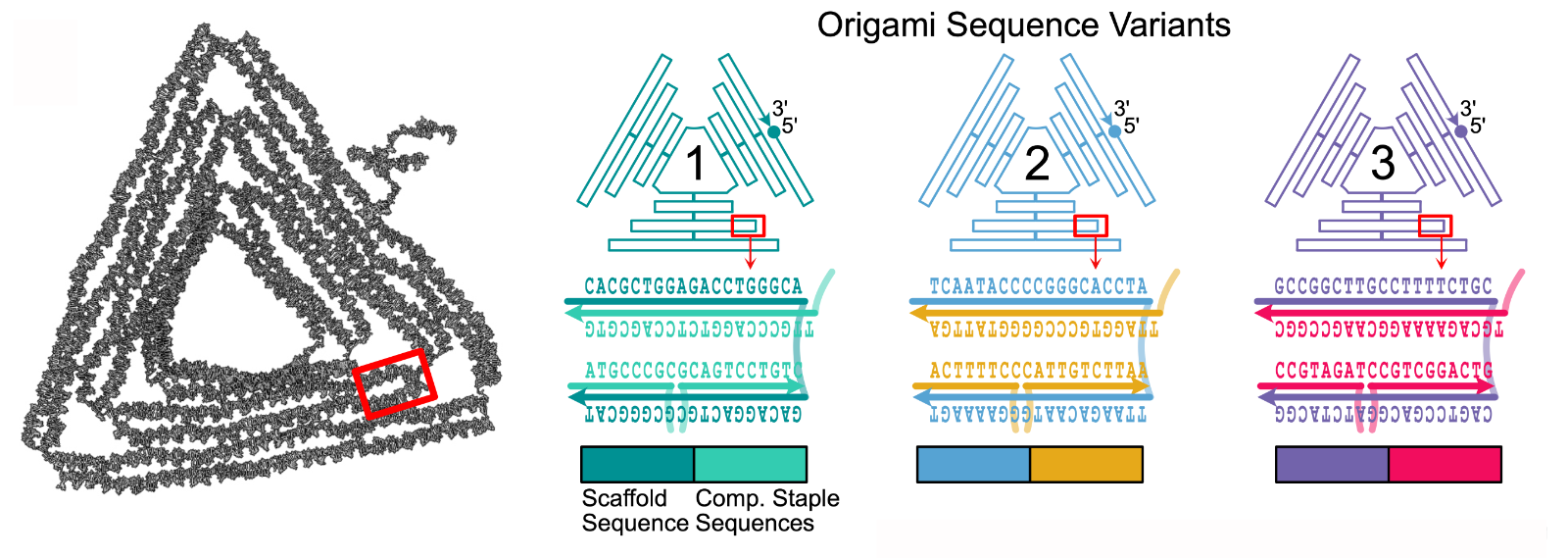
⬆️ The same DNA origami triangle can be made by many different sets of DNA sequences. But which ones to choose?
It seems a miracle that DNA origami nanostructures can self-assemble at all. Typically, hundreds of different short DNA strands are mixed together with a longer DNA scaffold strand in a salty reaction buffer and violà! - after a thermal cooling period, millions of precision nanostructures emerge in the solution (like the triangle shown above, about 60 nanometers across).
But can this self-assembly process be improved further? Often the yield of a DNA origami nanostructure is not even close to 100%.
In this work, we investigated how DNA origami assembly can be affected by the choice of DNA sequences used. And, we came from a specific new angle: we investigated the effect of off-target reactions.
The A,C,G,T sequences for a DNA origami actually make possible a whole host of unintended (or off-target) binding sites, just down to chance complementarity between some sequence regions. For example, staple strands can sometimes bind to the wrong parts of the scaffold strand, or the scaffold strand can bind to itself in complex knots. Despite natural error correcting mechanisms like DNA strand displacement operating, we hypothesised that DNA origami self-assembly could be derailed by a sufficiently unlucky choice of sequences that gave rise to many off-target reactions.
All current software to design DNA origami nanostructures (as of 2025) does not pay attention to the off-target bindings that arise in the self-assembly process. Instead, it assumes that the correct on-target bindings between staples and scaffold will eventually form and win out. Therefore, in such software, sequence design is treated as just an afterthought, and the main focus is instead on drawing a correct geometric design compatible with the structural properties of DNA helices.
Conversely, in this work, we systematically considered off-target reactions arising from sequence choice. We developed a computational pipeline that determined where (e.g. what region of a biological plasmid) it is best to obtain the sequence of the scaffold strand such that off-target reactions during DNA origami self-assembly are minimised, given a specific DNA origami nanostructure design.
We pursued a novel approach whereby four types of off-target reaction were minimised simultaneously (a so-called a "multi-objective" approach). Our software ran an intensive calculation considering all binding sites and produced a web page of results, detailing which scaffold sequences are best to use for a particular DNA origami design.
To test our computational predictions, we extensively assembled triangle and rectangle 2D origamis, and tetrahedron and 6-helix bundle 3D origamis in the lab, making each shape several times but with different complementary DNA sequences each time.
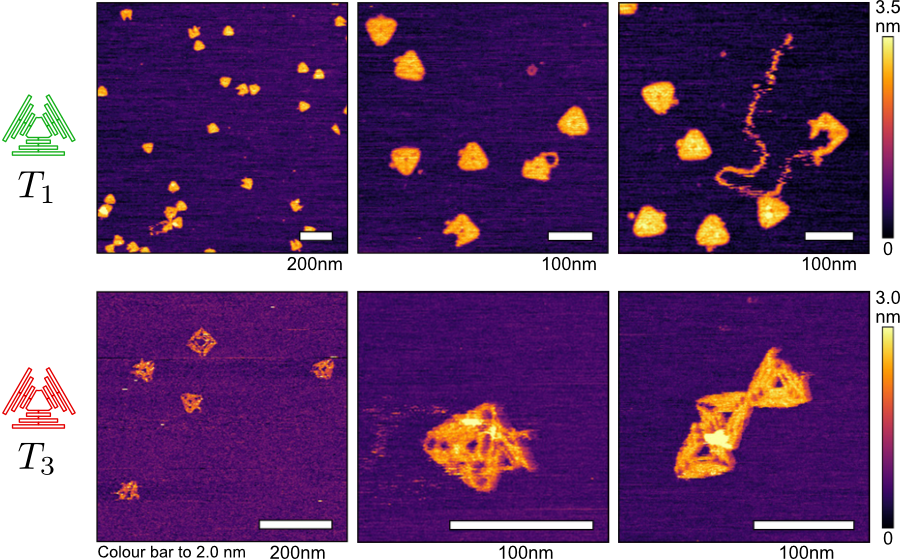
⬆️ A small DNA origami triangle imaged under AFM. Triangle T1 has a scaffold sequence causing minimal off-target reactions and assembles better than triangle T3 whose scaffold sequence causes many off-target reactions.
For the four origami shapes we assembled we found that, indeed, it was possible to make unfortunate choices for sequences. We found that "bad" sequence regions of the Lambda DNA vector (that create many off-target reactions) do exist, and lead to poor origami assembly in all cases when selected as the scaffold sequence. Conversely, we could use our software to see what region of the Lambda DNA vector (or any other biological vector, like a plasmid) is optimal for creating these DNA origami designs.
We hope this work can further contribute to the creation of precision nanometer-scale self-assembling structures.
Journal Paper
Shirt-Ediss and Torelli et al. (2025). Optimizing DNA origami assembly through selection of scaffold sequences that minimise off-target interactions. BioRxiv preprint. Currently under revision in Nature Communications.
Supplementary Material for the paper.
How to Install and Use
https://scaffoldselector.readthedocs.io
🤖 Helper tools:
Software Repositories
https://bitbucket.org/engineering-data-structure-organoids/scaffoldselector (License: Academic Non-commercial License)
🤖 Helper tools:
- https://bitbucket.org/engineering-data-structure-organoids/contactmap
- https://bitbucket.org/engineering-data-structure-organoids/poolgen
Techniques Used
Multi-objective selection, Pareto front, multi-criteria decision making (MCDM), DNA thermodynamics, approximated fast-compute DNA energy model, reaction site enumeration, HPC computing, HTML reporting
This work was developed while I was a senior postdoctoral researcher at Newcastle University, UK.



