REVNANO: From DNA Sequences to DNA Origami Designs
DNA Nanotech Posted 23 August 2025
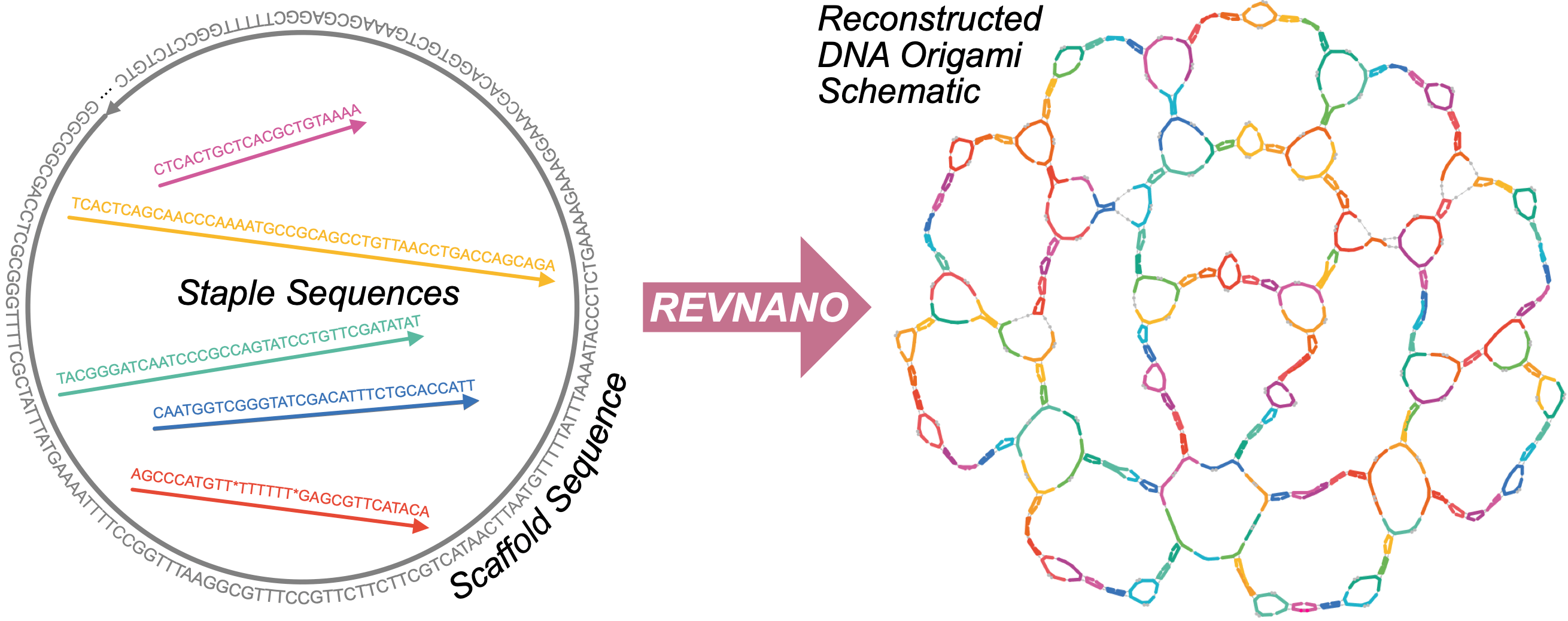
REVNANO attempts to solve an interesting reverse engineering problem in DNA nanotechnology; a challenge that has not been tackled until now!
When labs fabricate a DNA origami object, they first use some specialised design software to draw the object in 2D or 3D. When the design is completed, the software outputs the complementary DNA strand sequences needed to make the object self-assemble at the nanoscale. The DNA strands (a long "scaffold" strand, and hundreds of smaller "staple" strands) are then ordered and mixed together in the lab, making billions of the origami nano-object in just a drop of reaction buffer.
However, when it comes to publishing their work, labs often often omit the original electronic design files for their DNA origami and include just the raw DNA sequences they used. And publishing just the raw sequences can be a real problem for others who want to check the sequences actually work, or who want to modify the DNA origami design for their own purposes: all they can see are long lists of letters A,C,G and T.
REVNANO is a software program which tries to go the opposite way: it takes the published sequences for a DNA origami, and from them, it tries to re-construct a visual version of the original DNA origami design file (as an interactive web page). Users can then inspect the design and re-purpose it for their own needs.
The smiley face DNA origami design below was re-generated just from its DNA sequences; a task that would be near impossible for a human!
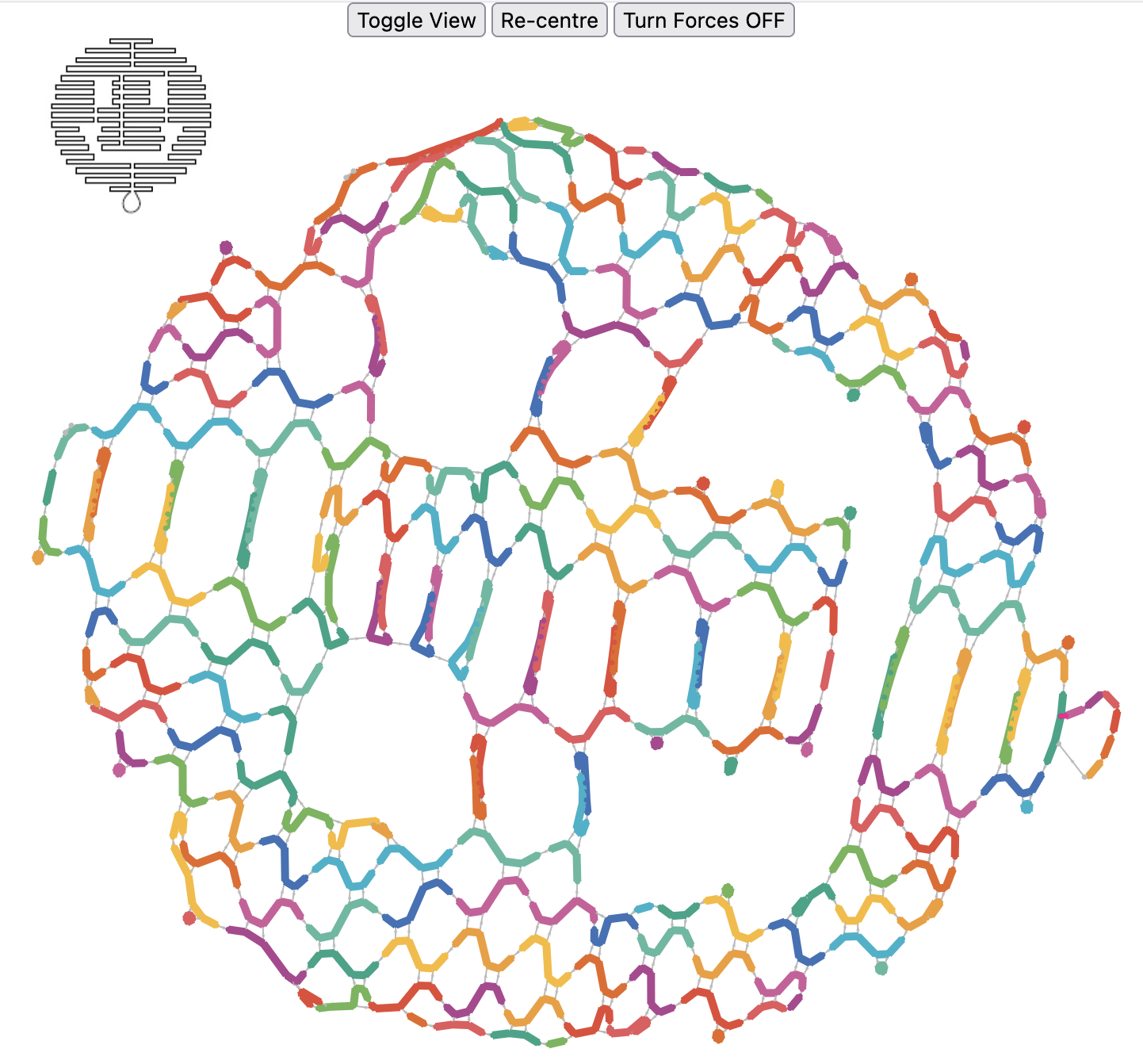
⬆️ The Rothemund smiley face DNA origami (top left) reverse engineered from its sequences. Click the image for the interactive web page, where the view can be changed to see the scaffold strand routing, the staple strand routings, or to see which junctions can possibly move (red) due to chance sequence complementarity.
This type of "reverse problem" is known as a constraint satisfaction problem (CSP) in Computer Science, and it's in the same general problem class as solving a Sudoku puzzle or completing a jigsaw. To address the problem, REVNANO implemented a custom CSP solver, and then used a clever graph layout algorithm (called the Kamada–Kawai algorithm) to draw the design in 2D or 3D space.
REVNANO was tested on 36 diverse origami designs from the literature and found that 29 origamis (81 %) had a good quality guide schematic recovered from raw DNA sequences!
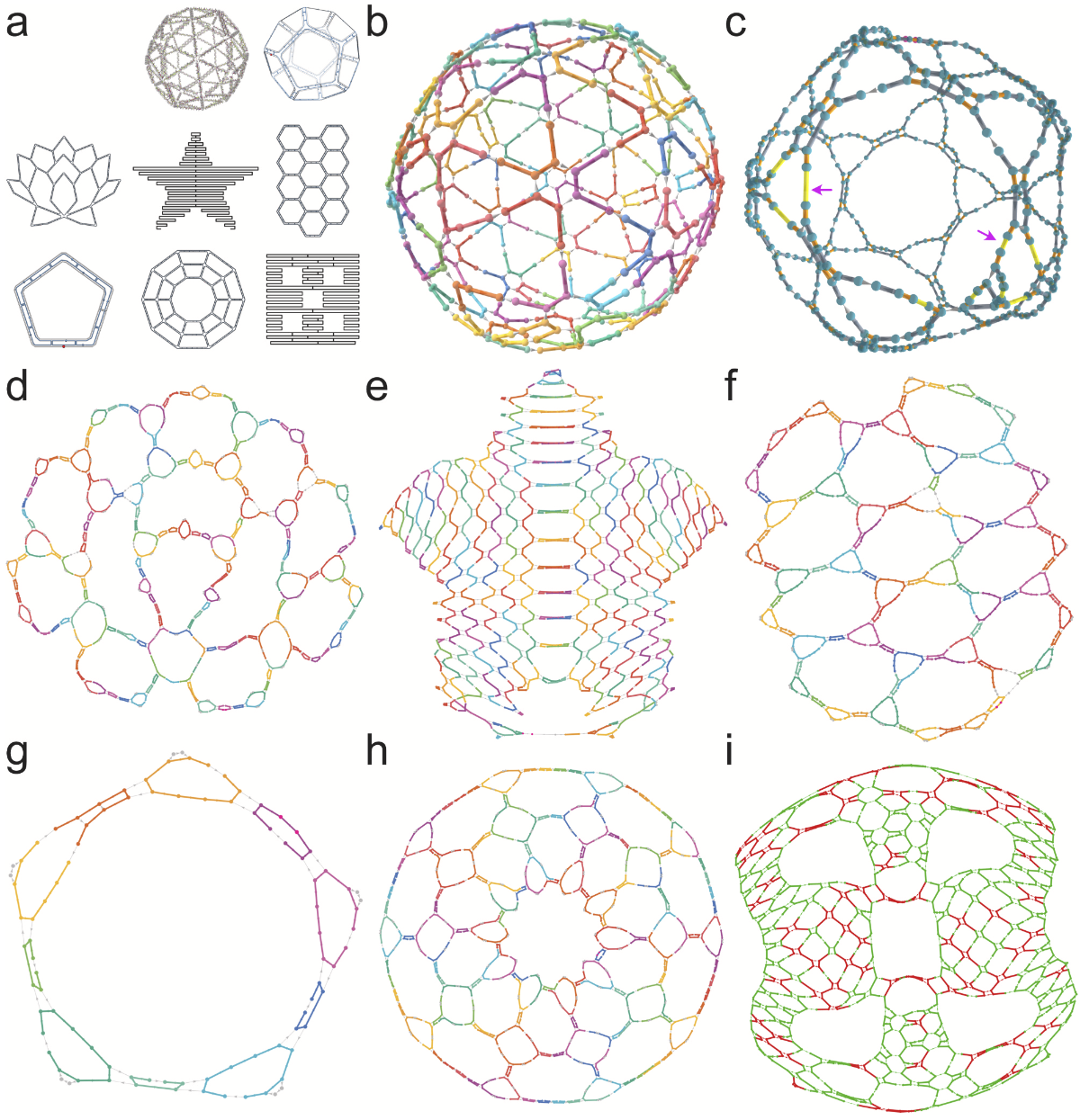
⬆️ Eight different DNA origami designs reverse engineered from their DNA sequences with REVNANO. Click the image for an interactive web page showing the 3D Ball in (b), reverse engineered from these sequences.
Journal Paper
Shirt-Ediss et al. (2023). Reverse engineering DNA origami nanostructure designs from raw scaffold and staple sequence lists. Computational and Structural Biotechnology 21, 3615-3626
Supplementary Material for the paper.
Presentation Slides
Slides for an invited talk I gave at Edinburgh University, October 2023. ©2023 Ben Shirt-Ediss.
How to Install and Use
https://revnano.readthedocs.io
Software Repository
https://bitbucket.org/engineering-data-structure-organoids/revnano (License: MIT License)
Techniques Used
Custom constraint satisfaction solver, Tree generation/pruning algorithms, Spring embedder, Kamada–Kawai algorithm, Physics engine, HPC computing, 3D JavaScript rendering
This work was developed while I was a senior postdoctoral researcher at Newcastle University, UK.



