DNA Stack: Storing Data in a Chemical Memory Made of DNA Polymers
DNA Nanotech Posted 24 August 2025
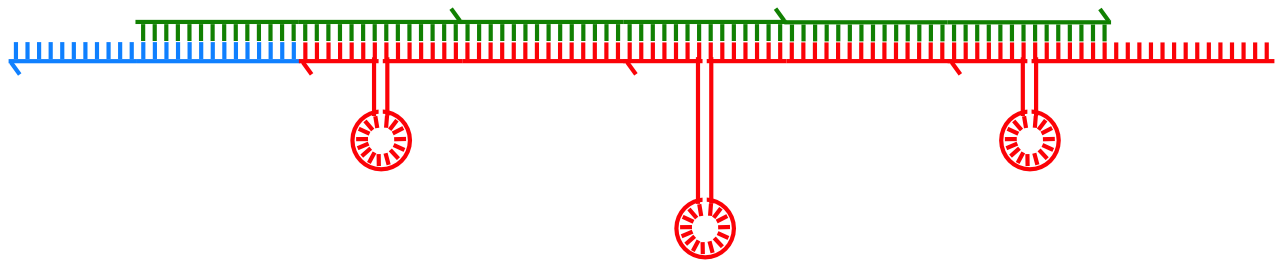
This project involved both experimental lab and computational modelling components working in parallel, to see if we could implement a simple memory in a test tube by using DNA strands.
The memory had a particular property: what went in last came out first (a so-called "stack" memory). Instead of bits, the memory held DNA strands of different types. Adding a bit, or performing an operation, involved adding a certain DNA strand type to the reaction. After a "bit" was read from the memory, the strand type representing that bit increased in concentration in the test tube.
The underlying chemistry was a DNA polymerisation reaction, where single strands would combine to form longer polymers as information was added. Then, the DNA polymers would become truncated shorter again as the information was read out.
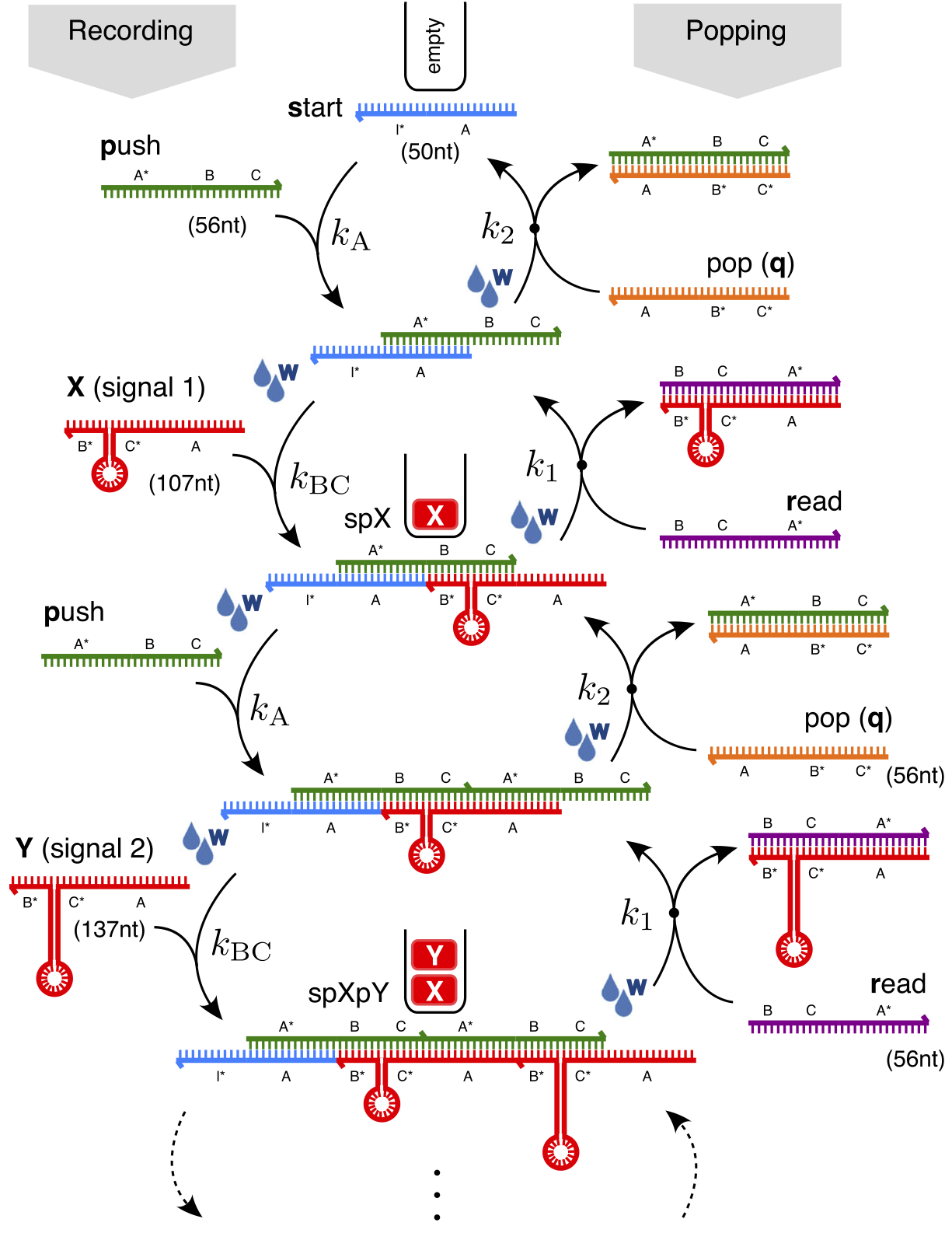
⬆️ Operation of the DNA stack memory, recording information XY (going down) and then 'popping' it back off again in reverse order (going up). Different coloured lines represent DNA strands with different sequences.
The actual reaction happening in the test tube was much more complicated than the idealised description in the image above, and so to try to understand it, we developed a computational kinetics model of the chemistry using rule-based modelling. In this type of modelling, you don't specify all possible reactions beforehand, instead you specify more abstract matching rules which add/remove reactions from the model only as certain products come into existence/disappear.
Further, the experimental results we got were often noisy and varied day to day. Therefore, we calibrated the model parameters to experimental data by using a novel technique we developed called intensity orderings. Under this new technique, the model scored well if it could reproduce just the relative order of intensities of gel bands in the experimental results (rather than their absolute intensities, which could vary considerably between runs of the same experiment).
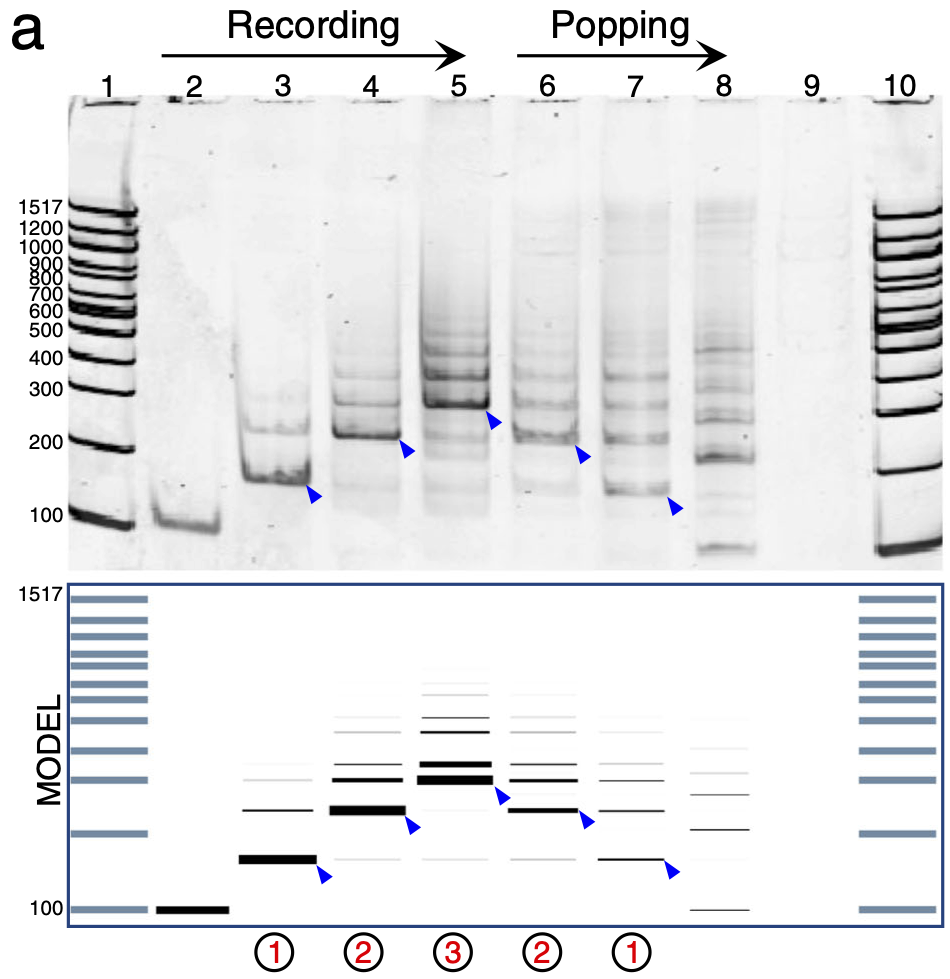
⬆️ Experimental results (top) vs. computational modelling results (bottom), as the DNA stack is used to record three bits, and then pop them off in reverse order.
The calibrated model agreed well with the experimental observations, and we went on to use just the model as a pure exploration tool, to see what the limits of the DNA chemical memory were without having to do thousands of experiments.
We quantified how much the "washing" procedure (performed in the lab in between DNA stack operations) would have to improve, in order to record more bits in the memory. Under our lab protocol, we were limited to recording 3 to 5 bits. Also we quantified how the memory device would respond to different sequences of recording and popping signals.
Journal Paper
Lopiccolo and Shirt-Ediss et al. (2021). A last-in first-out stack data structure implemented in DNA. Nature Communications 12, 4861
Supplementary Material for the paper.
How to Install and Use DNA Stack Model
https://dnastack.readthedocs.ioSoftware Repository
https://bitbucket.org/engineering-data-structure-organoids/dnastack (License: MIT License)Techniques Used
DNA chemical kinetics modelling, Gillespie algorithm, rule-based stochastic simulation, volume awareness, novel parameter estimation from experiments, kinetic rate constant fitting from UV data, HPC computing
This work was developed while I was a postdoctoral researcher at Newcastle University, UK.



